QGT Analysis Plan - 2021
Hae Kyung Im
2021-06-22
Last updated: 2021-06-30
Checks: 6 1
Knit directory: QGT-Columbia-lab/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200603) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version dee90ff. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: extras/.DS_Store
Unstaged changes:
Modified: analysis/2021_analysis_plan.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/2021_analysis_plan.Rmd) and HTML (docs/2021_analysis_plan.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | a1eed46 | Hae Kyung Im | 2021-06-30 | uploaded slides |
| html | a1eed46 | Hae Kyung Im | 2021-06-30 | uploaded slides |
| Rmd | 97ee478 | Hae Kyung Im | 2021-06-24 | no need to run comment |
| html | 97ee478 | Hae Kyung Im | 2021-06-24 | no need to run comment |
| html | 69ecf8e | Hae Kyung Im | 2021-06-22 | knitted |
| Rmd | 1a6b399 | Hae Kyung Im | 2021-06-22 | analysis plan 2021 |
| html | 1a6b399 | Hae Kyung Im | 2021-06-22 | analysis plan 2021 |
| Rmd | b47937d | Hae Kyung Im | 2021-06-22 | analysis plan todo |
| html | b47937d | Hae Kyung Im | 2021-06-22 | analysis plan todo |
| Rmd | 383c29f | Hae Kyung Im | 2021-06-22 | renaming by year |
| html | 383c29f | Hae Kyung Im | 2021-06-22 | renaming by year |
Here we will perform some of the analyses we discussed in the lecture this morning to gain hands on experience. Follow the steps listed below to get your RStudio environment1.
Getting access to the Rstudio environment
- Find your Rstudio server IP address and get the username and password here
- connect to the Rstudio server using the url you claimed (http://xxx.xxx.xxx.xxx:8787) using a web browser
- log into the server using the username and password of the server you claimed
Summary of analysis plan
- predict whole blood expression
- check how well the prediction works with GEUVADIS expression data
- run association between predicted expression and a simulated phenotype
- calculate association between expression levels and coronary artery disease risk using s-predixcan
- fine-map the coronary artery disease gwas results using torus
- calculate colocalization probability using fastenloc
- run transcriptome-wide mendelian randomization in one locus of interestgi
Initial remarks
- We ask you to actively participate in today’s hands on activities. Notice that we may ask you to share your screen for pedagogic purposes.
- If you have any concerns about this, please ask me or one of the TAs for assistance. We are here to help you learn.
- As you run the analysis and programs, we ask you to find the tab with your name and respond the questions in this document. Find the tab with your name and fill out the questions as you go along.
- You are welcome to check other people’s answers as guidelines but please make sure you write down your own answers.
Preliminary definitions
- Go to the terminal tab on the RStudio server and run the following commands to update the analysis document to the most recent version.
## Copy the lines below using Ctrl-C (or Cmd-C), paste on the terminal, and hit enter
PRE="/home/student/"
cd $PRE/lab/
git pull Under the Files tab (right bottom panel of RStudio) navigate to the folder Home/lab/analysis and open 2021_analysis_plan.Rmd (click on the file name)
activate the the imlabtools environment, which will make sure all the necessary python modules are available to the software we will be running.
## Copy the lines below using Ctrl-C (or Cmd-C), paste on the terminal, and hit enter
conda activate imlabtoolsReminder: the bash chunks need to be copy-pasted to the terminal, not performed within the chunk.
- execute the following chunk to load the tidyverse package, a convenient set of R packages for analysis
suppressPackageStartupMessages(library(tidyverse))- To define some variables to access the data more easily within the R session, run the following r chunk
print(getwd())
lab="/home/student/lab"
CODE=glue::glue("{lab}/code")
source(glue::glue("{CODE}/load_data_functions.R"))
source(glue::glue("{CODE}/plotting_utils_functions.R"))
PRE="/home/student/QGT-Columbia-HKI"
MODEL=glue::glue("{PRE}/models")
DATA=glue::glue("{PRE}/data")
RESULTS=glue::glue("{PRE}/results")
METAXCAN=glue::glue("{PRE}/repos/MetaXcan-master/software")
FASTENLOC=glue::glue("{PRE}/repos/fastenloc-master")
TORUS=glue::glue("{PRE}/repos/torus-master")
TWMR=glue::glue("{PRE}/repos/TWMR-master")
# This is a reference table we'll use a lot throughout the lab. It contains information about the genes.
gencode_df = load_gencode_df()- define some variables to access the data more easily in the terminal. Remember we are running R code in the R console and command line code in the terminal.
## Copy the lines below using Ctrl-C (or Cmd-C), paste on the terminal, and hit enter
export PRE="/home/student/QGT-Columbia-HKI"
export LAB="/home/student/lab"
export CODE=$LAB/code
export DATA=$PRE/data
export MODEL=$PRE/models
export RESULTS=$PRE/results
export METAXCAN=$PRE/repos/MetaXcan-master/software
export TWMR=$PRE/repos/TWMR-masterTranscriptome-wide association methods
Now we will perform a transcriptome-wide association analysis using the PrediXcan suite of tools. 
We start by predicting the expression levels of genes using the genotype data and the prediction weights and then perform an association between the predicted expression and the phenotype (denoted trait in the figure below). 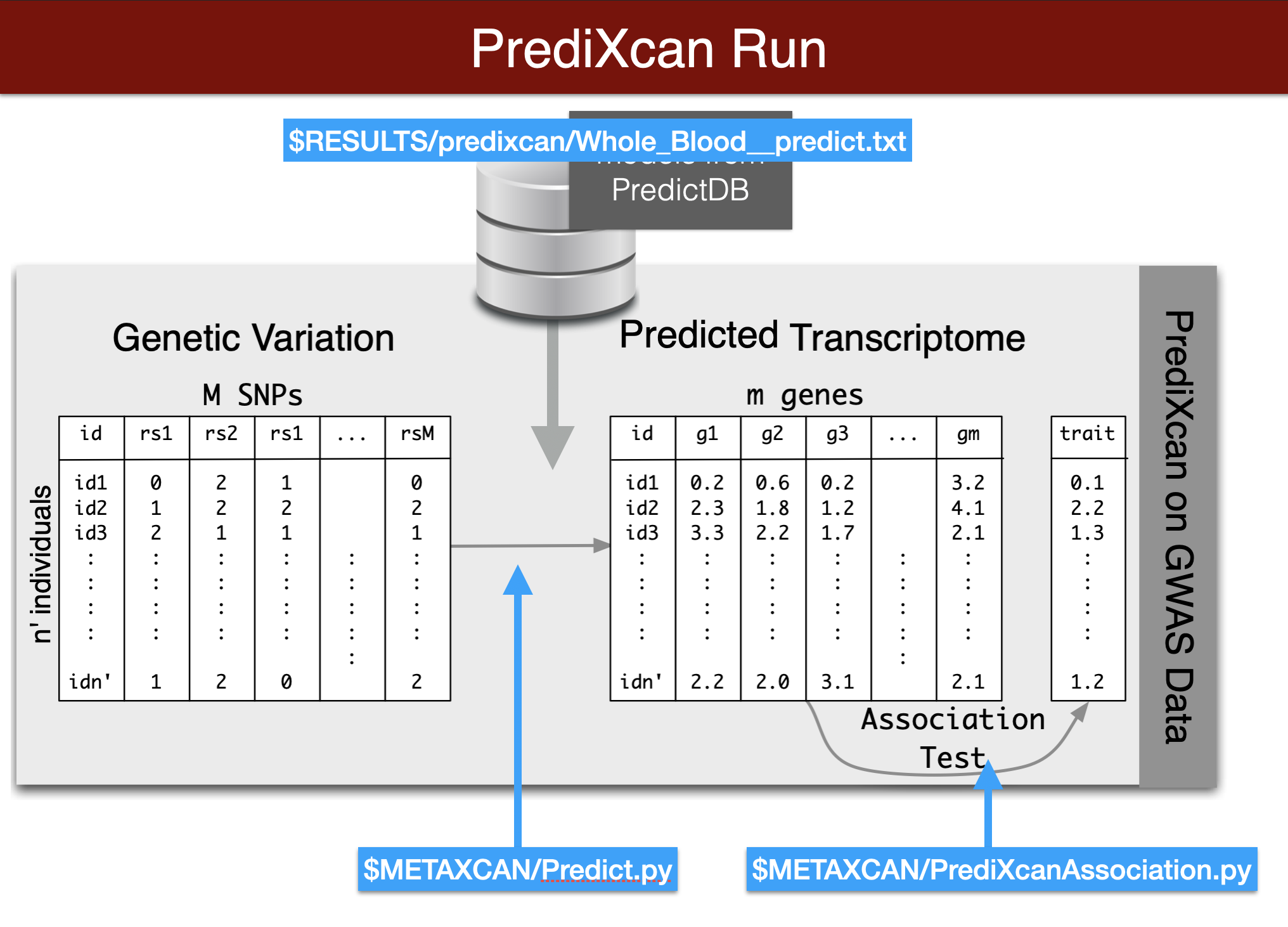
predict expression
We will predict expression of genes in whole blood using the Predict.py code in the METAXCAN folder.
Prediction models (weights) are located in the MODEL folder. Additional models for different tissues and transcriptome studies can be downloaded from predictdb.org.
Remember you need to copy and paste this code chunk into the terminal to run it. Also make sure you activated the imlabtools environment which has all the necessary python modules.
Make sure all the paths and file names are correct
This run should take about one minute.
run the following code in the terminal.
## Copy the lines below using Ctrl-C (or Cmd-C), paste on the terminal, and hit enter
printf "Predict expression\n\n"
python3 $METAXCAN/Predict.py \
--model_db_path $PRE/models/gtex_v8_en/en_Whole_Blood.db \
--vcf_genotypes $DATA/predixcan/genotype/filtered.vcf.gz \
--vcf_mode genotyped \
--variant_mapping $DATA/predixcan/gtex_v8_eur_filtered_maf0.01_monoallelic_variants.txt.gz id rsid \
--on_the_fly_mapping METADATA "chr{}_{}_{}_{}_b38" \
--prediction_output $RESULTS/predixcan/Whole_Blood__predict.txt \
--prediction_summary_output $RESULTS/predixcan/Whole_Blood__summary.txt \
--verbosity 9 \
--throw
- run following code in the console to get information on reported prediction performance.
prediction_fp = glue::glue("{RESULTS}/predixcan/Whole_Blood__predict.txt")
## Read the Predict.py output into a dataframe. This function reorganizes the data and adds gene names.
predicted_expression = load_predicted_expression(prediction_fp, gencode_df)
head(predicted_expression)
## read summary of prediction, number of SNPs per gene, cross validated prediction performance
prediction_summary = load_prediction_summary(glue::glue("{RESULTS}/predixcan/Whole_Blood__summary.txt"), gencode_df)
## number of genes with a prediction model
dim(prediction_summary)
head(prediction_summary)
print("distribution of prediction performance r2")
summary(prediction_summary$pred_perf_r2)assess actual prediction performance (optional)
## download and read observed expression data from GEUVADIS
## from https://uchicago.box.com/s/4y7xle5l0pnq9d1fwmthe2ewhogrnlrv
## Remove the version number from the gene_id's (ENSG000XXX.ver)
head(predicted_expression)
## merge predicted expression with observed expression data (by IID and gene)
## plot observes vs predicted expressioni for
## ERAP1 (ENSG00000164307)
## PEX6 (ENSG00000124587)
## calculate spearman correlation for all genes
## what's the best performing gene?run association with a simulated phenotype
\(Y = \sum_k T_k \beta_k + \epsilon\)
with random effects \(\beta_k \sim (1-\pi)\cdot \delta_0 + \pi\cdot N(0,1)\)
export PHENO="sim.spike_n_slab_0.01_pve0.1"
printf "association\n\n"
python3 $METAXCAN/PrediXcanAssociation.py \
--expression_file $RESULTS/predixcan/Whole_Blood__predict.txt \
--input_phenos_file $DATA/predixcan/phenotype/$PHENO.txt \
--input_phenos_column pheno \
--output $RESULTS/predixcan/$PHENO/Whole_Blood__association.txt \
--verbosity 9 \
--throw
More predicted phenotypes can be found in $DATA/predixcan/phenotype/. The naming of the phenotypes provides information about the genic architecture: the number after pve is the proportion of variance of Y explained by the genetic component of expression. The number after spike_n_slab represents the probability that a gene is causal \(\pi\)(i.e. prob \(\beta \ne 0\))
read association results
## read association results
PHENO="sim.spike_n_slab_0.01_pve0.1"
predixcan_association = load_predixcan_association(glue::glue("{RESULTS}/predixcan/{PHENO}/Whole_Blood__association.txt"), gencode_df)
## take a look at the results
dim(predixcan_association)
predixcan_association %>% arrange(pvalue) %>% head
predixcan_association %>% arrange(pvalue) %>% ggplot(aes(pvalue)) + geom_histogram(bins=20)
## compare distribution against the null (uniform)
gg_qqplot(predixcan_association$pvalue, max_yval = 40)compare estimated effects with true effect sizes
truebetas = load_truebetas(glue::glue("{DATA}/predixcan/phenotype/gene-effects/{PHENO}.txt"), gencode_df)
betas = (predixcan_association %>%
inner_join(truebetas,by=c("gene"="gene_id")) %>%
select(c('estimated_beta'='effect',
'true_beta'='effect_size',
'pvalue',
'gene_id'='gene',
'gene_name'='gene_name.x',
'region_id'='region_id.x')))
betas %>% arrange(pvalue) %>% head
## do you see examples of potential LD contamination?
betas %>% ggplot(aes(estimated_beta, true_beta))+geom_point()+geom_abline()Summary PrediXcan
Now we will use the summary results from a GWAS of coronary artery disease to calculate the association between the genetic component of the expression of genes and coronary artery disease risk. We will use the SPrediXcan.py.
The GWAS results (harmonized and imputed) for coronary artery disease are available in $PRE/spredixcan/data/
run s-predixcan
python $METAXCAN/SPrediXcan.py \
--gwas_file $DATA/spredixcan/imputed_CARDIoGRAM_C4D_CAD_ADDITIVE.txt.gz \
--snp_column panel_variant_id --effect_allele_column effect_allele --non_effect_allele_column non_effect_allele --zscore_column zscore \
--model_db_path $MODEL/gtex_v8_mashr/mashr_Whole_Blood.db \
--covariance $MODEL/gtex_v8_mashr/mashr_Whole_Blood.txt.gz \
--keep_non_rsid --additional_output --model_db_snp_key varID \
--throw \
--output_file $RESULTS/spredixcan/eqtl/CARDIoGRAM_C4D_CAD_ADDITIVE__PM__Whole_Blood.csv
plot and interpret s-predixcan results
spredixcan_association = load_spredixcan_association(glue::glue("{RESULTS}/spredixcan/eqtl/CARDIoGRAM_C4D_CAD_ADDITIVE__PM__Whole_Blood.csv"), gencode_df)
dim(spredixcan_association)
spredixcan_association %>% arrange(pvalue) %>% head
spredixcan_association %>% arrange(pvalue) %>% ggplot(aes(pvalue)) + geom_histogram(bins=20)
gg_qqplot(spredixcan_association$pvalue)- SORT1, considered to be a causal gene for LDL cholesterol and as a consequence of coronary artery disease, is not found here. Why? (tissue)
Exercise
run s-predixcan with liver model, do you find SORT1? Is it significant?
compare zscores in liver and whole blood.
run multixcan (optional)
- multixcan aggregates information across multiple tissues to boost the power to detect association. It was developed movivated by the fact that eQTLs are shared across multiple tissues, i.e. many genetic variants that regulate expression are common across tissues.
python $METAXCAN/SMulTiXcan.py \
--models_folder $MODEL/gtex_v8_mashr \
--models_name_pattern "mashr_(.*).db" \
--snp_covariance $MODEL/gtex_v8_expression_mashr_snp_smultixcan_covariance.txt.gz \
--metaxcan_folder $RESULTS/spredixcan/eqtl/ \
--metaxcan_filter "CARDIoGRAM_C4D_CAD_ADDITIVE__PM__(.*).csv" \
--metaxcan_file_name_parse_pattern "(.*)__PM__(.*).csv" \
--gwas_file $DATA/spredixcan/imputed_CARDIoGRAM_C4D_CAD_ADDITIVE.txt.gz \
--snp_column panel_variant_id --effect_allele_column effect_allele --non_effect_allele_column non_effect_allele --zscore_column zscore --keep_non_rsid --model_db_snp_key varID \
--cutoff_condition_number 30 \
--verbosity 7 \
--throw \
--output $RESULTS/smultixcan/eqtl/CARDIoGRAM_C4D_CAD_ADDITIVE_smultixcan.txt
Colocalization methods
- Colocalization methods seek to estimate the probability that the complex trait and expression causal variants are the same. We favor methods that calculate the probability of causality for each trait (posterior inclusion probability), called fine-mapping methods. Here we use torus for fine-mapping and fastENLOC for colocalization.
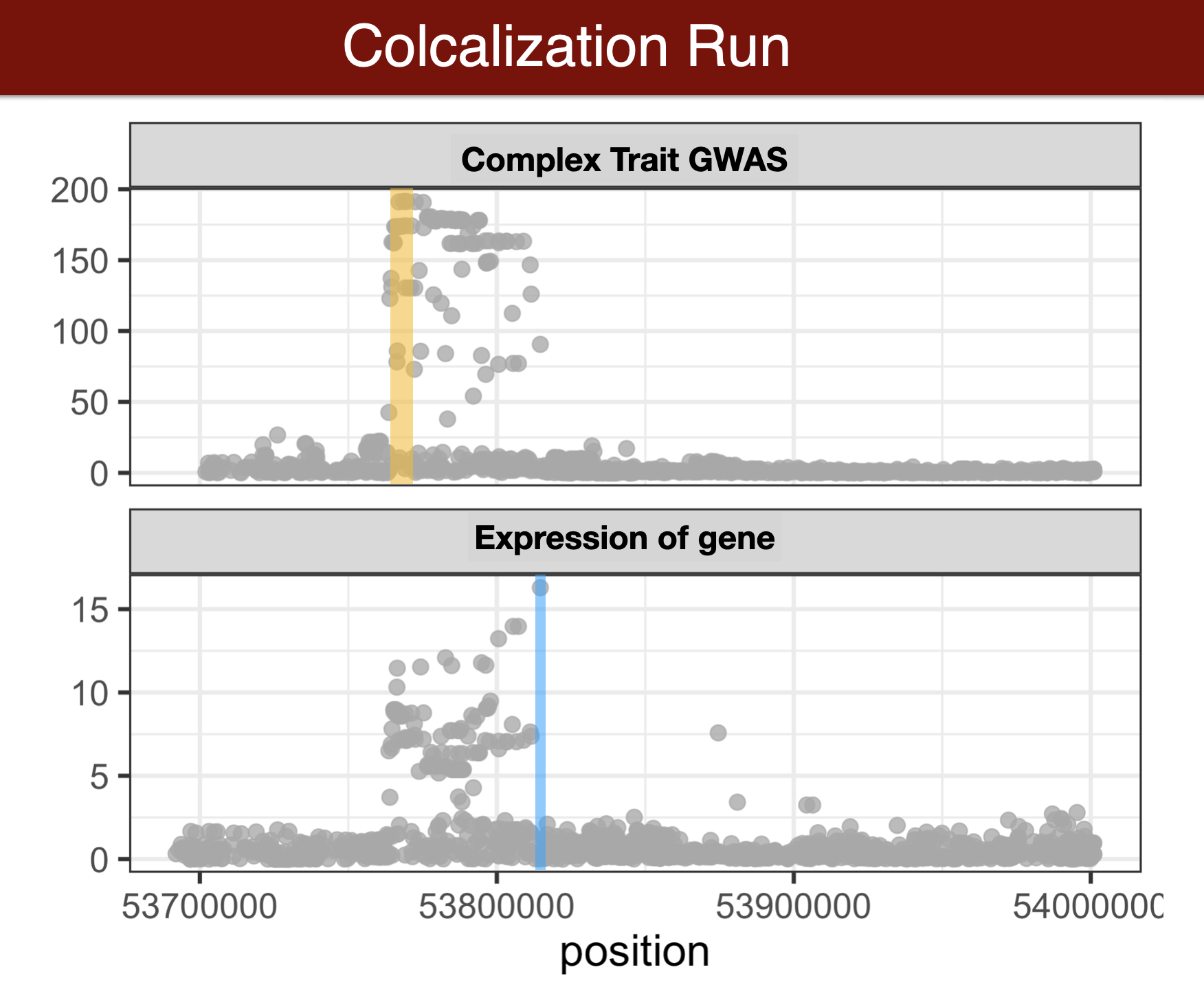
GWAS summary statistics to torus format
the following code will format GWAS summary statistics into a format that the fine-mapping method torus can understand.
we precalculated this for you so there is no need to run the following chunk
## THERE IS NO NEED TO RUN THIS CHUNK
## We ran this formatting for you because it takes over 10 minutes.
python $CODE/gwas_to_torus_zscore.py \
-input_gwas $DATA/spredixcan/imputed_CARDIoGRAM_C4D_CAD_ADDITIVE.txt.gz \
-input_ld_regions $DATA/spredixcan/eur_ld_hg38.txt.gz \
-output_fp $DATA/fastenloc/CARDIoGRAM_C4D_CAD_ADDITIVE.zval.gzfine-map GWAS results
We run torus due to time limitation but ideally we would like to run a method that allows multiple causal variants per locus, such as DAP-G or SusieR.
torus has been precompiled and placed within the PATH
export TORUSOFT=torus
$TORUSOFT -d $PRE/data/fastenloc/CARDIoGRAM_C4D_CAD_ADDITIVE.zval.gz --load_zval -dump_pip $PRE/data/fastenloc/CARDIoGRAM_C4D_CAD_ADDITIVE.gwas.pip
cd $PRE/data/fastenloc
gzip CARDIoGRAM_C4D_CAD_ADDITIVE.gwas.pip
cd $PRE We can take a quick look at the z-values and finemapping PIPs (posterior probability of causality):
cd $PRE/data/fastenloc
zless CARDIoGRAM_C4D_CAD_ADDITIVE.zval.gz
zless CARDIoGRAM_C4D_CAD_ADDITIVE.gwas.pip.gzcalculate colocalization with fastENLOC
## you can take a look at the tutorial https://github.com/xqwen/fastenloc/tree/master/tutorial
export eqtl_annotation_gzipped=$PRE/data/fastenloc/FASTENLOC-gtex_v8.eqtl_annot.vcf.gz
export gwas_data_gzipped=$PRE/data/fastenloc/CARDIoGRAM_C4D_CAD_ADDITIVE.gwas.pip.gz
export TISSUE=Whole_Blood
export FASTENLOCSOFT=fastenloc
##export FASTENLOCSOFT=/Users/owenmelia/projects/finemapping_bin/src/fastenloc/src/fastenloc
mkdir $RESULTS/fastenloc/
cd $RESULTS/fastenloc/
$FASTENLOCSOFT -eqtl $eqtl_annotation_gzipped -gwas $gwas_data_gzipped -t $TISSUE
#[-total_variants total_snp] [-thread n] [-prefix prefix_name] [-s shrinkage]
analyze results
## optional - compare with s-predixcan results
fastenloc_results = load_fastenloc_coloc_result(glue::glue("{RESULTS}/fastenloc/enloc.sig.out"))
spredixcan_and_fastenloc = inner_join(spredixcan_association, fastenloc_results, by=c('gene'='Signal'))
ggplot(spredixcan_and_fastenloc, aes(RCP, -log10(pvalue))) + geom_point()
## which genes are both colocalized (rcp>0.10) and significantly associated (pvalue<0.05/number of tests)Mendelian randomization methods
run TWMR (for a locus)
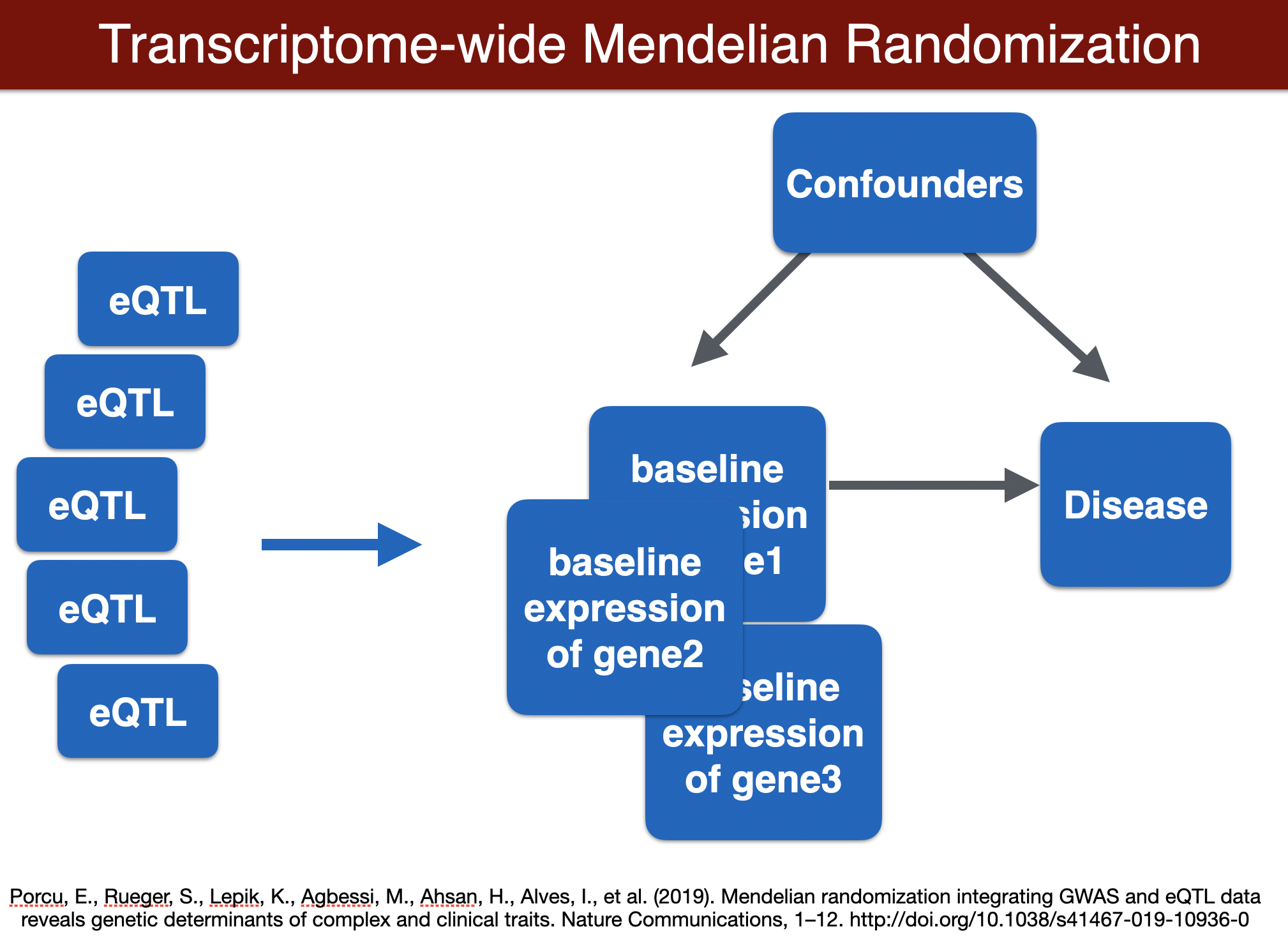
TWMR
# Load the 'analyseMR' function
source(glue::glue("{CODE}/TWMR_script.R"))
# Collect the list of genes available to run
gene_lst <- list.files(TWMR)
gene_lst <- gene_lst[str_detect(gene_lst, "ENS.*")]
gene_lst <- (gsub("\\..*", "", gene_lst) %>% unique)
# Set the gene and run. The function writes output to a file.
for (gene in gene_lst) {
analyseMR(gene, TWMR)
}twmr_results <- load_twmr_results(TWMR, gencode_df)Optional items
Setting up your own system
Linux is the operating system of choice to run bioinformatics software. You will need either a computer running linux or or mac os, which has a linux-like environment.
- install anaconda/miniconda
- define imlabtools conda environment how to here, which will install all the python modules needed for this analysis session
- download data and software from Box. This will have copies of all the software repositories and the models
-
download software
- download metaxcan repo
- download torus repo
- download fastenloc repo
- download TMWR repo
- download prediction models from predictdb.org
- install R/RStudio/tidyverse package
- (optional) install workflowr package in R
- git clone https://github.com/hakyimlab/QGT-Columbia-HKI.git
- start Rstudio (if you installed workflowr, you can just open the QGT-Columbia-HKI.Rproj)
How to create RStudio server
Check instructions here
Slides
Download slides here
Acknowledgements
Contributions by Owen Melia, Yanyu Liang, and Tyson Miller
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Catalina 10.15.7
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] Rcpp_1.0.6 whisker_0.4 knitr_1.30 magrittr_2.0.1
[5] workflowr_1.6.2 R6_2.5.0 rlang_0.4.10 fansi_0.4.2
[9] stringr_1.4.0 tools_4.0.3 xfun_0.20 utf8_1.2.1
[13] git2r_0.28.0 htmltools_0.5.1 ellipsis_0.3.1 rprojroot_2.0.2
[17] yaml_2.2.1 digest_0.6.27 tibble_3.1.0 lifecycle_1.0.0
[21] crayon_1.4.1 later_1.1.0.1 vctrs_0.3.6 promises_1.1.1
[25] fs_1.5.0 glue_1.4.2 evaluate_0.14 rmarkdown_2.6
[29] stringi_1.5.3 compiler_4.0.3 pillar_1.5.1 httpuv_1.5.5
[33] pkgconfig_2.0.3note that these servers are different from the Rstudio cloud you used yesterday↩︎